The job of molecular biologists – the ones who discover things like leptin and AMPK and superoxide dismutase – is to figure out the genetics and enzyme pathways behind it all.
I’ve worked as a molecular biologist. It’s admittedly been awhile, but I can tell you that they’re very interested in the interactions between enzymes in signalling networks. It can be very easy to get lost in that forest. It is easy to forget the simple statement that a Cornell professor said to me that I’ve always remembered, “All biology is evolutionary biology.” As a budding molecular biologist – interested in all things to do with DNA and RNA and how it worked – this didn’t really hit home at the time. I mean sure, all of these pathway must have EVOLVED at some point.
But I wanted to take the clock apart and see how it WORKED! Look at which gear was driving what shaft.
But you can run into problems here because biology tends to be a bit messy. You spend seven years doing a thesis about the molecular interactions of NF-kB and the year after you publish your results someone points out that NF-kB is actually activated by being acetylated. Whoops. Important point, you probably wished you’d known that! Et cetera.
Molecular biologists are nerds and they spend most of their lives in the weeds of what proteins cause other proteins to be produced, which other proteins those proteins interact with once they are produced and how they communicate with each other: phosphorylation, methylation, acetylation, ubiquination, lipoylation, etc.
In my opinion, it would make sense for them to come up out of the weeds more, pull back and think about the big picture from an evolutionary perspective. I’m interested in energy balance. I think about evolution. I drew this onion.
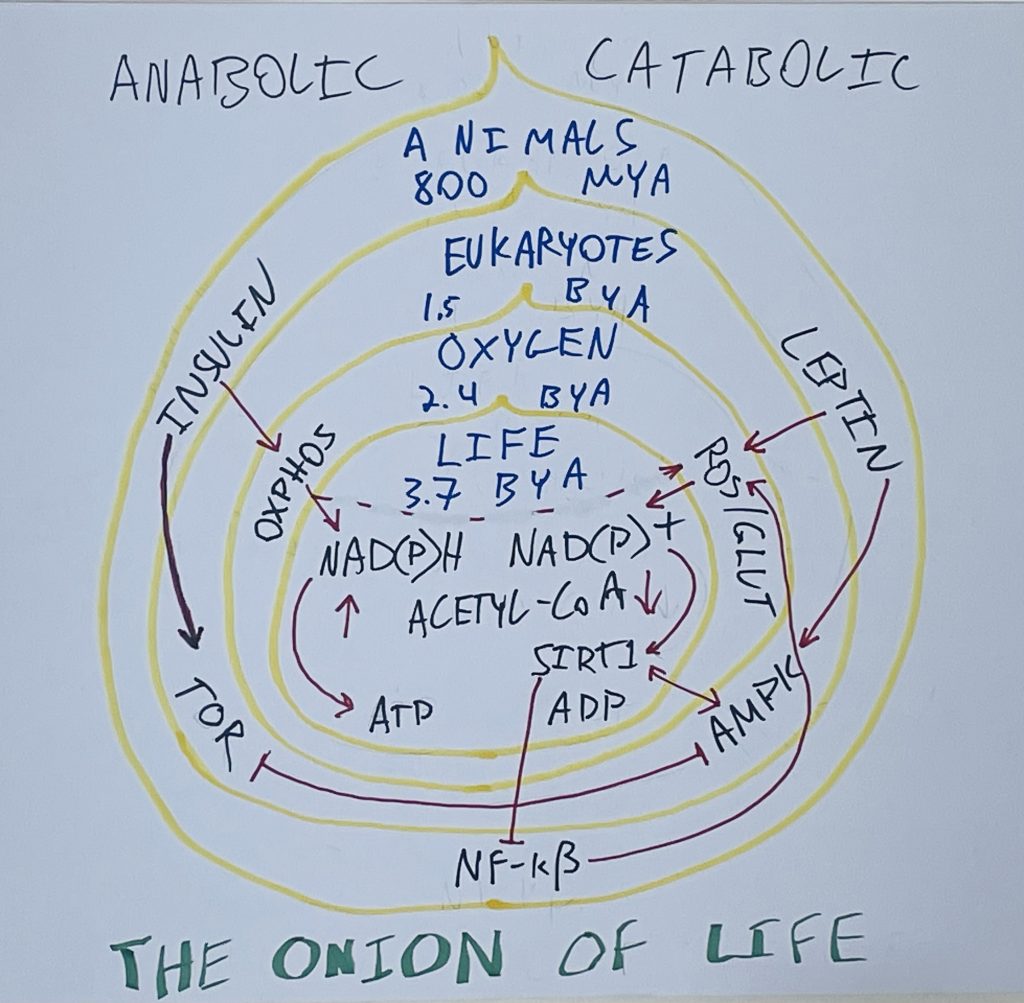
Please send photos of how much better your sixth grader’s art is than mine.
At the core of the onion are the things relevant to energy balance that are shared by all living cells. These things evolved 3.7 BILLION years ago. NADH and NAD+. NADPH and NADP+. Acetyl-CoA. Deacetylase enzymes that rely on NAD+ and acetyl-CoA levels. ATP and ADP. You could throw in the process of glycolysis here if you were so inclined.
These things are 3.7 billion years old and these are the things that control whether or not cells are anabolic (building) or catabolic (breaking down). This is how ALL cells work. Bacterial cells, plant cells, the cells of archaea; ameoba cells – a single celled eukaryote; and your cells. When NADH, ATP and acetyl-CoA levels are high, the cell is in anabolic mode. What a cell chooses to do when in anabolic mode is context dependent. An E. coli cell (a bacteria) in anabolic mode will reproduce its DNA and replicate. A fat cell in anabolic mode will store fat.
Video Version
If you prefer to watch/listen:
Oxygen
Sometime before 2.4 billion years ago cyanobacteria evolved the enzyme systems needed to use the power of the sun to make glucose from carbon dioxide. They excreted molecular oxygen as a waste product. By 2.4 billion years ago oxygen levels were rising in the atmosphere, putting most life on earth into a crisis. Oxygen is corrosive. It causes rust and fire and at the time that the oxygenated atmosphere came about the vast majority of organisms on earth couldn’t live in its presence. Many still cannot: we call these obligate anaerobes.
The things that evolved to live in the newly oxygenated atmosphere evolved robust antioxidant defense systems: superoxide dismutase; glutathione, glutathione peroxidase and glutathione reductase; thioredoxin, thioredoxin peroxidase and thioredoxin reductase; catalase; quinols.
Oxygen also became the terminal electron acceptor in oxidative phosphorylation systems involving electron transport chains. This allowed cells to use the energized electrons in a molecule of acetyl-CoA to convert three molecules of NAD+ to NADH. NADH is the reduced form of NAD that carries the electrons. OILRIG: oxidation is losing, reduction is gaining (electrons). Electron transport chains and the Krebs cycle allowed us to extract MUCH more energy out of glucose as ATP.
NADH pushes the cell towards the anabolic side of the onion. For this reason, electron transport chains and Krebs cycle enzymes such as pyruvate dehydrogenase release a steady stream of Reactive Oxygen Species (ROS) as they work. Our antioxidant system removes the ROS. In this process NADPH is converted back to NADP+, pushing the balance of the cell back towards catabolic. ROS provide energetic balance.
Don’t Reinvent The Wheel
Here’s where the onion analogy comes into play. After the oxygen catastrophe there was a wave of innovation involving electron transport chains and antioxidant defense enzymes. But the core systems that informed cells whether or not they should divide and grow or conserve energy stayed the same. Those systems – involving NADH/NAD+ ratios, sirtuin enzymes and acetyl-CoA levels – remained intact. They already had 1.3 billion years to work out the kinks.
The oxygen enzymes were layered on top of the core life program. This is analogous to DOS. Bill Gates bought the DOS operating system and built Microsoft Windows on top of it. DOS is the part of the operating systems that knows how to run programs, write things to memory and the hard drive, to draw pixels on the screen. Windows is a Graphical User Interface (GUI). It allows people to start programs using a mouse. It’s convenient, but in a Windows system DOS is still running the computer. Windows talks to DOS when it wants it to write to the hard drive and DOS lets Windows know that it’s written the thing to the hard drive. But DOS is running the hard drive.
Cells are the same. The oxygen enzymes evolved to manipulate the NADH/NAD+ and acetyl-CoA levels. In turn they are manipulated by the NADH/NAD+ and acetyl-CoA levels through processes such as acetylation and deacetylation. The cell is still making its core decisions based on the NADH/NAD+ and acetyl-CoA levels.
Eukaryotes
And so biology goes on like this. Layers are added. There are brief periods of intense evolution followed by long periods of relative stasis. The next true biological epoch after oxygen was the development of eukaryotes, which involved a cell without an electron transport chain engulfing one that did. The engulfed cell came to live inside of the other cell – the first mitochondria.
This all happened by at least 1.5 billion years ago.
Eukaryotes have intracellular organelles. Nuclei, endoplasmic reticula, mitochondria, etc. Organelles require more signalling (communication). The eukaryotic layer of energy regulation brings into play AMPK and TOR, the yin and yang of energy signalling. AMPK is catabolic, activating sirtuin enzymes (Sirt1 in mammals) and being activated by sirtuin enzymes. TOR activates pathways in building fats and proteins and prevents autophagy: the breaking down and recycling of old organelles.
One way that AMPK achieves its catabolic role is by supressing the production of SCD1, an enzyme which converts saturated fats (SFA) to monounsaturated fats (MUFA). My next post will be the discussion of SCD1 and The Onion. MUFA create less ROS when burned in the mitochondria and ROS is catabolic. Conversely, AMPK is turned OFF by becoming acetylated, which happens when acetyl-CoA and NADH levels are high: when the cell is in anabolic mode.
The signalling molecules in the eukaryotic layer are doing their job by manipulating and being manipulated by the things happening in the core layer.
Animals
The layers are somewhat arbitrary. I could have had multi-cellular life, followed by animals, followed by vertebrates, followed by mammals. The big point here is that animals have different specialized tissues. Tissues require another level of communication. Insulin and leptin are hormones that circulate in the bloodstream that alert different tissues to organismal energy status.
Again, you don’t reinvent the wheel for cells to work in some new way due to insulin and leptin. Insulin signals TOR and leptin signals AMPK. The animal layer simply interacts with the next layer down. The cells are still running off of NADH/NAD+ and acetyl-CoA. Just like cells always have.
In specialized tissues, cells will behave differently when they are anabolic or catabolic based on the context around them. Muscle cells are not fat cells. All cells judge anabolic versus catabolic the same way, though.
Let’s Compare
I like the onion because it gives context to what is and what came before. The brief bursts of evolution put down new layers of regulation over what existed before. That is how biology works.
Let’s look at AMPK regulation from the perspective of a molecular biologist. This is taken from KEGG pathway. KEGG is a mainstream tool of molecular biologists.
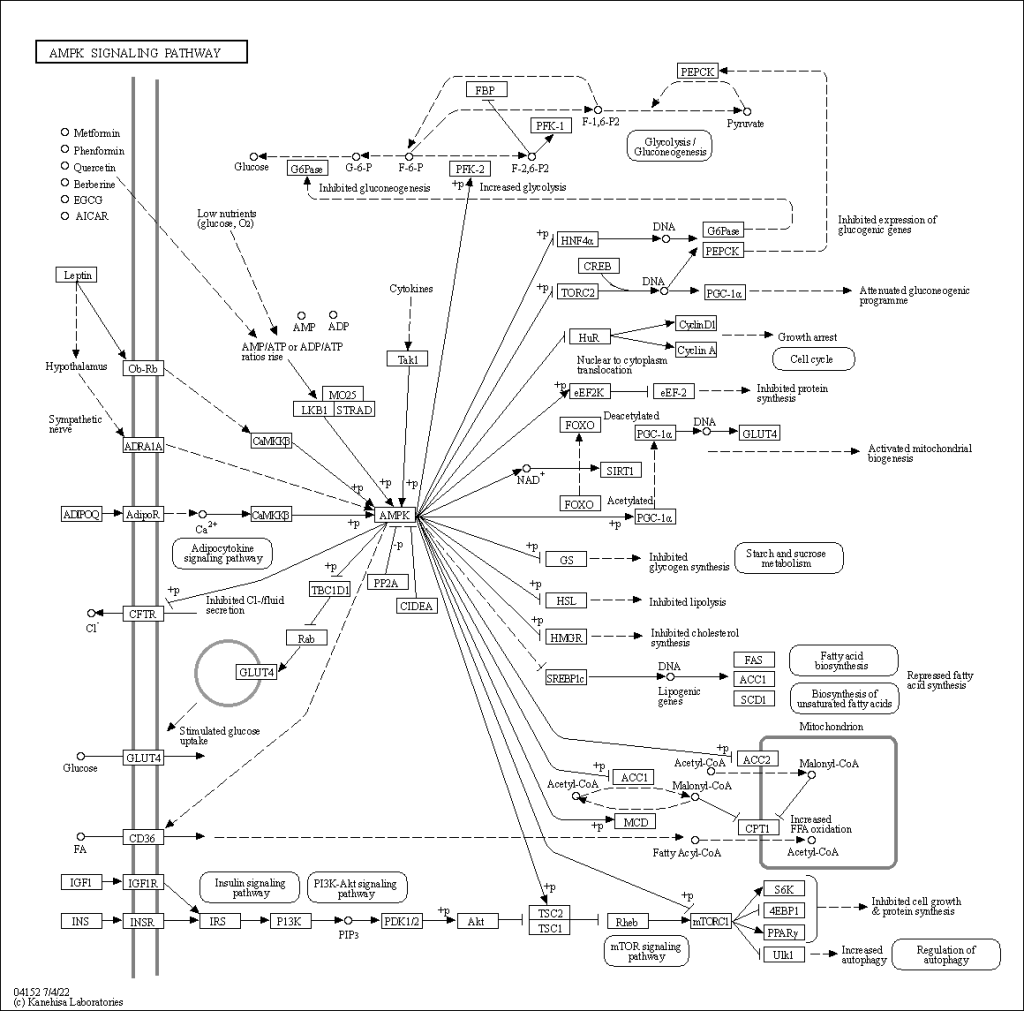
There’s no story here. There’s no context. There’s just a lot of things that interact with other things presented in a flat format. As we learn about more things that interact with other things we just add more things!
I don’t find this map particularly useful, so I made an onion.
Conclusion
All biology is evolutionary biology. What comes later does not replace what came before. It layers on top of it.
If you want to understand biology or why your body does the things it does, you have to start here. Start with the onion.
Check back for part two of this post. I’m going to move this conversation from the theoretical realm to the concrete one. I’m going to talk about my favorite enzyme again: SCD1, a crucial enzyme controlling fat storage. Almost 20 years went by before we started to get an idea of what controlled SCD1 levels, despite a hard decade of molecular biology. Hint: its whether the cell is anabolic or catabolic! In the meantime we found a lot of trees but missed the forest!
Roy A. Frye (2000). Phylogenetic Classification of Prokaryotic and Eukaryotic Sir2-like Proteins. , 273(2), 0–798. doi:10.1006/bbrc.2000.3000
Fischer, W. W., Hemp, J., & Valentine, J. S. (2016). How did life survive Earth’s great oxygenation? In Current Opinion in Chemical Biology (Vol. 31, pp. 166–178). Elsevier BV. https://doi.org/10.1016/j.cbpa.2016.03.013
Miller, A.-F. (2011). Superoxide dismutases: Ancient enzymes and new insights. In FEBS Letters (Vol. 586, Issue 5, pp. 585–595). Wiley. https://doi.org/10.1016/j.febslet.2011.10.048
Russell, M. J., & Martin, W. (2004). The rocky roots of the acetyl-CoA pathway. In Trends in Biochemical Sciences (Vol. 29, Issue 7, pp. 358–363). Elsevier BV. https://doi.org/10.1016/j.tibs.2004.05.007
Laurino, P., Tóth-Petróczy, Á., Meana-Pañeda, R., Lin, W., Truhlar, D. G., & Tawfik, D. S. (2016). An Ancient Fingerprint Indicates the Common Ancestry of Rossmann-Fold Enzymes Utilizing Different Ribose-Based Cofactors. In C. A. Orengo (Ed.), PLOS Biology (Vol. 14, Issue 3, p. e1002396). Public Library of Science (PLoS). https://doi.org/10.1371/journal.pbio.1002396

I am a former molecular biologist who left the field after more than a decade for exactly this reason – it’s very rare in the field for scientists to see the forest for the trees… I mean properly see or even want to see really. I thought the standard approach in the field led to huge amounts of waste of time and money and other resources. It is also why I was hooked by Peter’s early Protons posts all those years ago. It was such a zoomed out view compared to the molecular biology world and I thought it would be a better approach.
But, the inner mol biologist hasn’t gone away – I look at this KEGG pathway and think it’s fascinating! I’m looking forward to this series.
Some people (e.g. Nick Lane – Power, Sex Suicide) argue that the proton gradient is even involved in the origin of life.
Brad, I was about to order alpha lipoic acid from your shop, but I saw that there is no list of ingredients. It’s very important for me to know because my husband has multiple allergies. For example, we cannot use any of the vitamin shop store brands because nearly all of them contain rice flour.
I’m asking here because I couldn’t find any contact information anywhere. Thanks.
Haven’t checked back here in a little while but I like this post. Framing the underlying operating systems that stack on top of one another… which is a great analogy.